Based on mitogenome evidence, we found that the two Rhacophorus individuals we collected in Motuo County in the Tibet Autonomous Region, China were R. rhodopus (Figs. 5, 6 and 7). The two sequences have same gene order and both of them lose ND5gene, but one sequences (OK165559) is shorter than our reported sequences (OK181853). In our reported sequence, there are two more bases in the 16rRNA, and 37 more bases in D-loop. Here we only analyzed the structural characteristics of one sequences (OK181853).
Structural characteristics of the mitochondrial genome
We determined that the complete mitochondrial genome of R. rhodopus is 15,789 bp in length, and consists of 12 protein-coding genes (PCGs) (losing NADH dehydrogenase subunit 5), two ribosomal RNA genes (rRNAs), 22 transfer RNA genes (tRNAs), and a control region (D-loop) (Fig. 2 and Table S1). Among the 36 fragment genes, ND6 and eight genes (tRNA-Pro, tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser (UCN) and tRNA-Glu) were on the light strands (L-strand), and the remainder were located on the heavy strands (H-strand) (Fig. 2 and Table S1).
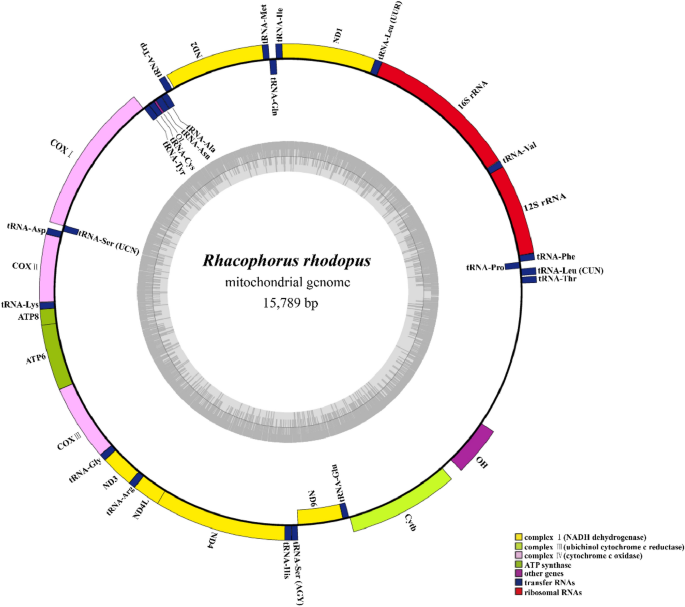
The complete mitochondrial genome sequence of Rhacophorus rhodopus collected from Motuo County in China.
The mitogenome structure of R. rhodopus was conserved, similar to the gene sequence structures of Rhacophorus schlegelii24 and Rhacophorus dennysi4. Four tRNA genes (tRNA-Thr, tRNA-Leu(CUN), tRNA-Pro and tRNA-Phe) formed a TLPF tRNA cluster, different from the neobatrachian-type arrangement25. In Rhacophoridae species, the ND5 gene is located between D-loop and tRNA-Thr4, but in R. rhodopus mtDNA, we found that the ND5 gene was lost. We also used the ND5 of other frog species to blast with R. rhodopus mtDNA and yet we did not find the ND5.The ND5 gene loss was also observed in Polypedates megacephalus9. This phenomenon may be common in vertebrates9,25.
The complete genome of R. rhodopus mtDNA consisted of 30.83% A, 30.03% T, 14.81% G, and 24.32% C. Similar to the base distribution in other anurans21, the A + T content (60.86%) was higher than the G + C content (39.14%), showing an obvious preference for A + T in the complete mitogenome sequences of this species. We also found that the AT-skew was 0.013 and the GC-skew was − 0.243, indicating more A than T, and more C than G (Table S2).
Protein-coding genes and codon usage patterns
Within the complete mitogenome genome of R. rhodopus, the total length of the 12 PCGs was 9,519 bp. Among the 12 PCGs, 11 PCGs (except COX I) used ATG as the initiation codon, while COX I genes initiated with an ATA codon. For COX I and ND6 the stop codon was AGG, whereas for ND3 and ND4L the stop codon was TAA. Seven protein genes (ATP6, COX II, COXIII, ND1, ND2, ND4 and Cytb) ended with incomplete stop codons TA- or T– (Table S2). These T–/TA- stop codons become a complete TAA stop codon through the post-transcriptional polyadenylation26.
The AT/CG-skews of the 12 PCGs are listed in Table S3. Except for ND2, ND6 and COX II, the remaining nine PCGs were negative in both the AT-skew and GC-skew. Meanwhile, the A + T content of the 12 PCGs was 60.37%; the AT-skew (− 0.074) and the GC-skew (− 0.253) were negative. The codon usage and relative synonymous codon usage (RSCU) values of R. rhodopus are shown in Table S3. Eight of 64 codons showed the highest use frequency and they were AUU (192), UUU (161), CUA (141), UUA (133), AUA (133), ACA (105), GCC (100), CUU (100). However, UCG and CGG codons were the least used stop codon.
Ribosomal RNA and transfer RNA genes
Similar to other vertebrates21, the mitogenome of R. rhodopus also included 12S and 16S rRNA genes, which were on the H strand. The 12S rRNA gene was 935 bp long and located between tRNA-Phe and tRNA-Val genes. The 16S rRNA gene with a length of 1,572 bp was located between tRNA-Val and tRNA-Leu (UUR) genes (Table S1). We found that the content of A + T (12S rRNA genes 54.55%, 16S rRNA genes 60.56%) was higher than that of C + G (12S rRNA genes 45.45%, 16S rRNA genes 39.44%). The AT-skew was slightly positive whereas the GC-skew was strongly negative (Table S2).
Of the 22 tRNA genes identified in the R. rhodopus mitogenome, 14 genes were located on the H strand and 8 genes were located on the L strand (Table S1). The secondary structure of tRNA is shown in Fig. 3. We found that 21 tRNA genes, except for tRNA-Ser (AGY), were able to form the classical cloverleaf secondary structure and that the use of anticodon was the same in other vertebrates3. In contrast, the tRNA-Ser (AGY) gene was unable to form a cloverleaf structure due to a lack of a dihydrouridine (DHU) arm. This is a common phenomenon in vertebrates27. However, Cheng et al.suggested that lack of DHU could become functional by adjusting its structural conformation to fit the ribosome in a similar way to that of usual tRNAs in the ribosomes28.
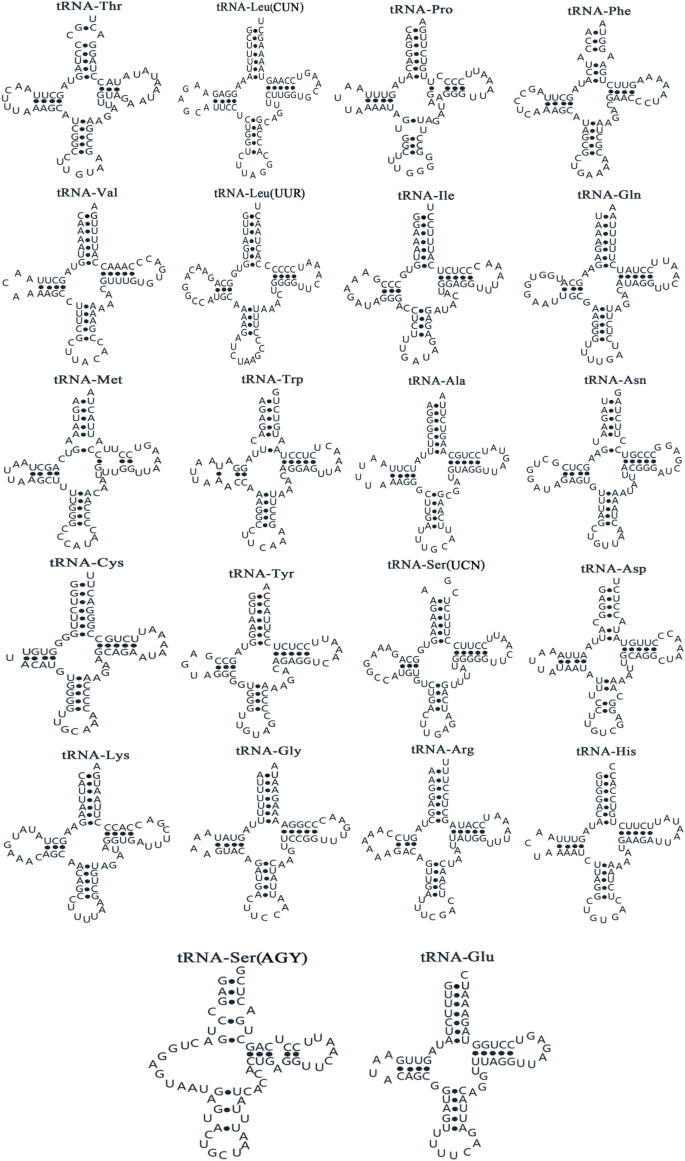
Putative tRNA secondary structures found mitochondrial genome of Rhacophorus rhodopus.
Noncoding regions
We identified two major noncoding regions in the R. rhodopus mitochondrial genome, at the origin of L-strand replication (OL) and in the control region (D-loop). The OL, a length of 23 bp, was located between tRNA-Asn and tRNA-Cys in the WANCY genes cluster (Table S1). The D-loop (2,230 bp), which was the longest part in the complete genome, was located between the Cytb gene and the tRNA-Thr gene on the H-strand (Table S1). Keddie et al. speculated that the D-loop may play an important role in gene replication29. In the D-loop sequence, A + T content was 67.17% and G + C content was 32.83%. In addition, AT/CG-skew analysis showed that the D-loop gene of R. rhodopus has a positive AT-skew (0.021), while the GC-skew (-0.180) was strongly negative (Table S2).
In general, the control regions contained several specific components, which can be easily identified by two tandem repeat units at two ends. We found that both 5′ and 3′-sides of the D-loop had two obvious repeat regions. One was 38 bp of 13.8 tandem repeat units (5′-TTGAAGGACA TACTATGTAT AATCACCATA TACTATGC-3′) on the 5′-side end, and the other was a 11 bp of 12.1 tandem repeat units (5′-TATATATGTAA-3′) on the 3′-side. In addition, three conserved sequence blocks (CSBs) were also detected (24 bp CSB-1, 5′-ATACCTGAAT GCTAGACGGA CATA-3′; 19 bp CSB-2, 5′-TACCCCCCCC TTTCCCCCC-3′; 17 bp CSB-3, 5′-CCTTAACACC CCCCCCG-3′) (Fig. 4 and Table S4). This phenomenon had also been observed in D-loops of other anuran species3,9.
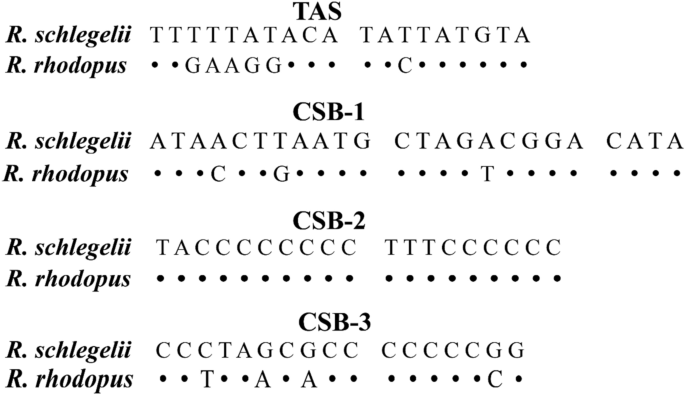
Structures and alignments of identified putative termination-associated sequences (TAS) and conserved sequence blocks (CSB1-3). Alignment gaps and nucleotides identical to the first line are indicated by a dot (·), respectively. Variable nucleotides are marked with corresponding nucleotides.
Phylogenetic relationships
We reconstructed BI and ML phylogenetic trees with three types of datasets, and both BI and ML phylogenetic trees showed similar topologies. Hence, we only show the BI tree in this study (Figs. 5, 6 and 7).
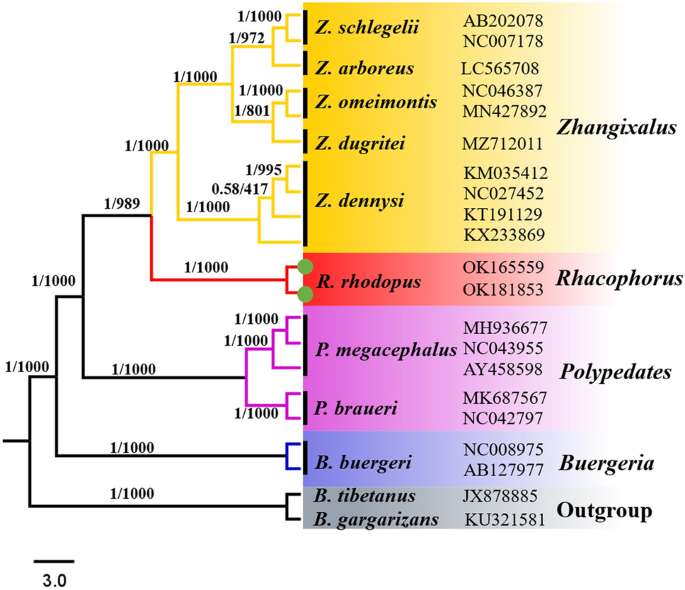
BI and ML analysis of 11 species complete mitochondrial genome sequence, Bufo gargarizans and Bufo tibetanus as outgroups. Tree topologies produced by BI and ML analyses were equivalent. Bayesian posterior probability (PP) and bootstrap support (BP) values for ML analyses are shown in order on the nodes.
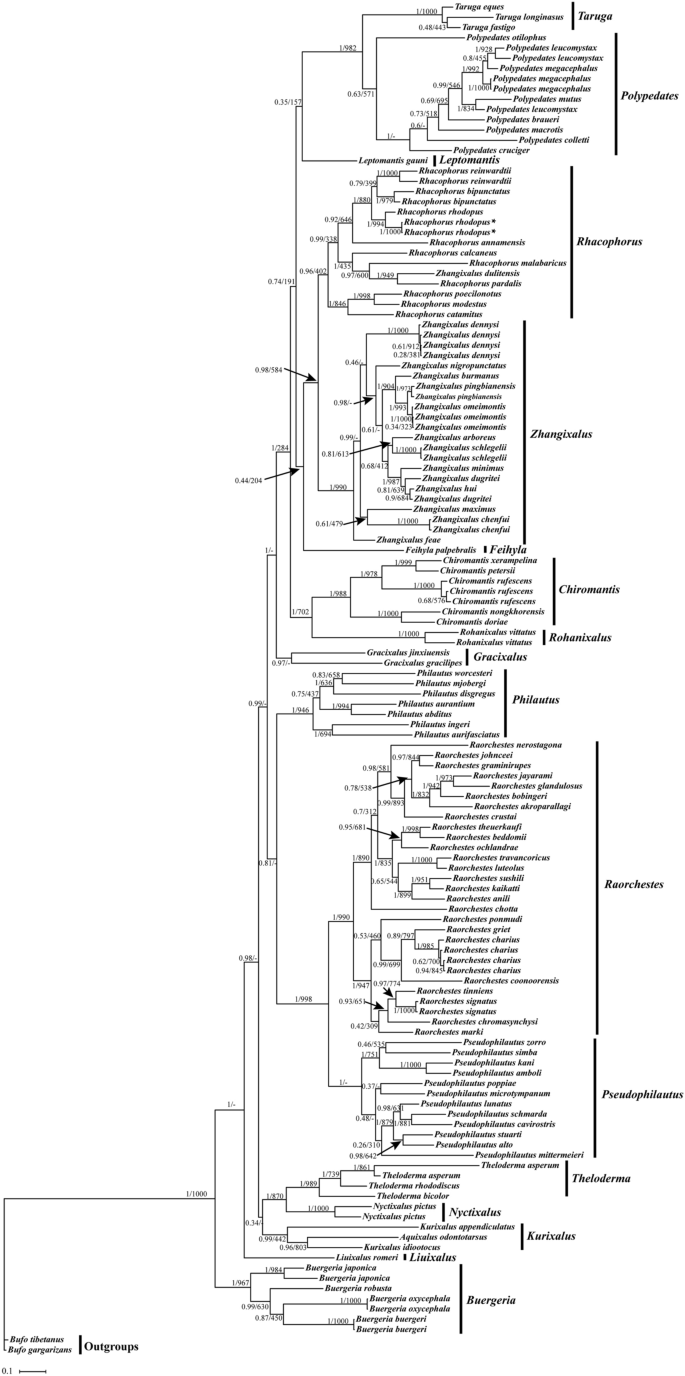
BI and ML analysis of 102 species based on 12S + 16S + CYTB genes sequence. Bufo gargarizans and Bufo tibetanus as outgroups. Tree topologies produced by BI and ML analyses were equivalent. Bayesian posterior probability (PP) and bootstrap support (BP) values for ML analyses are shown in order on the nodes.
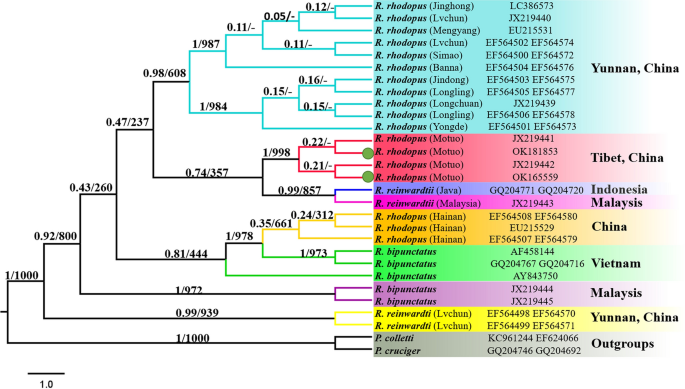
BI and ML analysis of Rhacophorus rhodopus, Rhacophorus bipunctatus and Rhacophorus reinwardtii, based on 12S and 16S. Polypedates colletti and Polypedates cruciger as outgroups. Tree topologies produced by BI and ML analyses were equivalent. Bayesian posterior probability (PP) and bootstrap support (BP) values for ML analyses are shown in order on the nodes.
Phylogenetic analysis based on the long mitochondrial genome data set
Our results show that Zhangixalus, Rhacophorus and Polypedates form a monophyletic group (PP = 1.00, BP = 1000), which supports the monophyletic origin of the tree frog family44. The phylogenetic tree is divided into two main branches. One separate branch is the genus Buergeria, the other main branch contains Zhangixalus, Rhacophorus and Polypedates (Fig. 5). Our results show that (Zhangixalus + Rhacophorus) is a sister clade of Polypedates (PP = 1.00, BP = 1000), which is consistent with a previous Rhacophoridae phylogenetic analyses4. In the Rhacophorus clade, two R. rhodopus from Tibet formed a clade with strong supports (PP = 1.00, BP = 1000) and the clade of R. rhodopus appeared as the sister taxon to the Z. dennysi + ((Z. omeimontis + Z. dugritei) + (Z. schlegelii + Z. arboreus)) clade (PP = 1.00, BP = 989). Here, Z. dennysi forms a monophyletic group, with strong support values (PP = 1.00, BP = 1000), which are similar to the results based on 16S rRNA gene phylogenetic analyses30.
Phylogenetic analysis based on the concatenated 12S + 16S + Cytb genes data set
We reconstructed BI and ML phylogenetic trees according to the concatenated 12S + 16S + Cytb gene dataset from 396 sequences of 104 Rhacophoridae species retrieved from NCBI (Fig. 6). Our results revealed monophyly of seven genera (Polypedates, Taruga, Kurixalus, Pseudophilautus, Theloderma, Raorchestes, and Buergeria) in the Rhacophoridae30. Previously, the genus Rhacophorus was divided into the genus Rhacophorus, Leptomantis and Zhangixalus31. However, in our study only Rhacophorus and Zhangixalus formed a clade. Furthermore, we found that R. pardalis and Z. dulitensis formed a sister group which was similar to the privious phylogenetic analysis of Meegaskumbura et al. and Chan et al.32,33. Our results also showed that R. bipunctatus and R. reinwardtii are more closely related than R. rhodopus. Thus, this result is consistent with Wilkinson et al.19, but does not support that R. bipunctatus was more closely related to R. rhodopus20,21.
Phylogenetic analysis of different populations of R. rhodopus based on the concatenated 12S and 16S rRNA genes dataset
To further explore the phylogenetic relationship of R. rhodopus in Tibet, we reconstructed BI and ML phylogenetic trees according to the concatenated 12S + 16S genes (842 bp) of R. rhodopus, R. reinwardtii and R. bipunctatus from different populations. The BI tree revealed that the three Rhacophorus individuals from the Motuo Tibet populations form a monophyletic clade, suggesting that they are the same species (R. rhodopus), which is consistent with the findings of Li et al.22. In addition, our results show that the two R. reinwardtii individuals from Yunnan China forms an independent branch, whereas R. reinwardtii from Indonesia and Malaysia, and the three R. rhodopus individuals from the Motuo Tibet populations also form a sister clade (Fig. 6). This suggests that R. reinwardtii may be paraphyletic34. Hence, the relationship among R. rhodopus, R. bipunctatus, and R. reinwardtii cannot be reliably solved and further research will explore the morphological features of R. reinwardtii and to reconfirm the accuracy of sequences of R. reinwardtii from Indonesia, Malaysia and from Lüchun County Yunnan Province of China.
The mitochondrial genome and phylogenetic analysis of Rhacophorus rhodopus & Latest News Update
The mitochondrial genome and phylogenetic analysis of Rhacophorus rhodopus & More Live News
All this news that I have made and shared for you people, you will like it very much and in it we keep bringing topics for you people like every time so that you keep getting news information like trending topics and you It is our goal to be able to get
all kinds of news without going through us so that we can reach you the latest and best news for free so that you can move ahead further by getting the information of that news together with you. Later on, we will continue
to give information about more today world news update types of latest news through posts on our website so that you always keep moving forward in that news and whatever kind of information will be there, it will definitely be conveyed to you people.
The mitochondrial genome and phylogenetic analysis of Rhacophorus rhodopus & More News Today
All this news that I have brought up to you or will be the most different and best news that you people are not going to get anywhere, along with the information Trending News, Breaking News, Health News, Science News, Sports News, Entertainment News, Technology News, Business News, World News of this made available to all of you so that you are always connected with the news, stay ahead in the matter and keep getting today news all types of news for free till today so that you can get the news by getting it. Always take two steps forward
Credit Goes To News Website – This Original Content Owner News Website . This Is Not My Content So If You Want To Read Original Content You Can Follow Below Links
