In silico ST diversity and population structure of K. pneumoniae in Portugal
This study includes a total of 509 Kp isolates. Of these, 459 are clinical isolates from hospitals in the southern (n = 378), central (n = 20) and northern (n = 61) regions of Portugal (Table 1). These were compared to isolates collected from veterinary clinics (n = 41) and environmental wastewater (n = 9) from the southern region, thereby broadening insights into the incidence of Kp in Portugal. The isolates were collected between years 1980 and 2019, but the majority (n = 455, 89%) were collected between 2000 and 2019 (Table 1). Of the O antigen types, O1 was dominant with O1v1 (44%) and O1v2 (20%) followed by O2v2 (12%) and O2v1 (10%) serotypes (Table 1). Seventy-seven different STs were inferred, with the most frequent being ST15 (n = 161), ST147 (n = 36), ST13 (n = 26) and ST14 (n = 26) (Table 1). Globally significant clonal group 258 had little presence, with 4% of isolates belonging to ST11 and none to ST258 or ST512. A high proportion of isolates (31%) were from low frequency STs (each < 2.0%). Out of 2,926 possible unique ST pairs, only 0.7% differed by a single allele, whereas 22.7%, 32.8% and 25.7% differed by 4, 5 and 6 alleles, respectively (Fig. S2). O serotypes were linked strongly with STs (Fig. S3). Only 10 STs had isolates with different O serotypes. ST15 (n = 161) had 149, 9 and 3 isolates with O1v1, O1v2 and O1/O2v1 serotypes, respectively. ST13 (n = 26) had 25 and 1 isolates with O1v2 and O1v1 serotypes, respectively. ST14 (n = 26) had 24 and 2 isolates with O1v1 and O1/O2v1 serotypes, respectively. ST348 (n = 22) had 19 and 3 isolates with O1v1 and O1/O2v1 serotypes, respectively. ST11 (n = 19) had 13, 5 and 1 isolates with O2v2, O3b and O4 serotypes, respectively. There were a further five STs (34 isolates) containing isolates with different O serotypes. The predominant capsular K serotypes were KL112 (14%) and KL24 (19%), linked to the common ST15. When considering only the unique ST and K group combinations (n = 105), the most frequent K serotypes were KL30, KL10, and KL24 observed in 7, 7 and 5 STs, respectively.
Phylogenetic analysis
The overall phylogenetic tree (n = 509) is largely congruent with ST type, but the propensity of Kp to recombine, means that the branch support values decline rapidly as one moves from leaves to root (Fig. S3). Individual ST trees (Fig. 1A–D) identified several well supported geographic clades, including two previously reported8,14 potential outbreaks in Lisbon and Vila Real hospitals involving representatives of ST147 and ST15. For the most abundant sequence type ST15 (n = 161), the tree shows multiple clades differentiated by K serotypes (Fig. 1A). Nearly all isolates carried blaCTX-M-15, except isolates from years 1980 to 1982 and 5 of 14 isolates from years 2007 to 2018 with blaKPC-3. Due to the low number of isolates spanning years 1990 to 1999, our dataset may not fully reflect the beta-lactamase diversity during that period. It has been suggested that blaTEM-10 was the dominant ESBL gene at that time, and most of our isolates (11/16) carried it. The smallest clade (n = 9) was from north Portugal and was distinguished by KL19 and O1v2 serotypes with five isolates carrying blaKPC-3. The other blaKPC-3 carrying ST15 isolates (n = 9) were phylogenetically very distant and part of a larger clade (n = 67) with K112 and O1v1 serotypes. Interestingly, these nine isolates carried both blaKPC-3 and blaCTX-M-15, in contrast to all other relevant isolates, which carry either blaKPC-3 or blaCTX- M-15 (Fig. 2). Four of the nine wastewater samples belonged to ST15, and were dispersed amongst clinical samples in the ST15 phylogenetic tree (Fig. 1A).
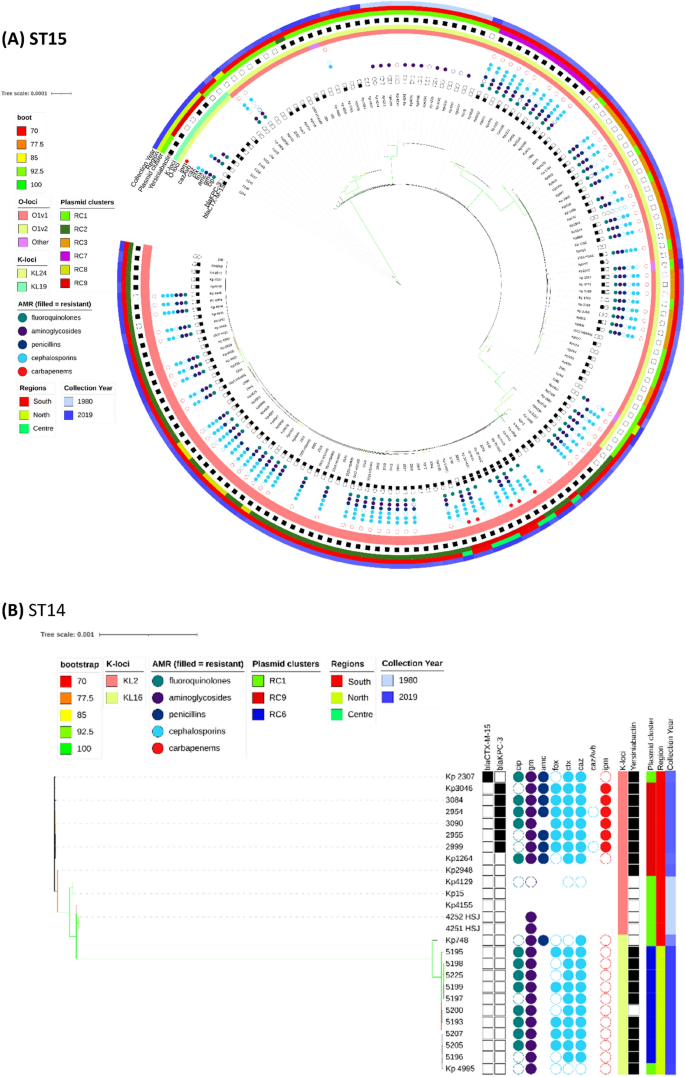
Phylogenetic trees of the most commons sequence types (STs), their antimicrobial resistance (AMR) phenotype, and carbapenemase and ESBL genotypic profiles. K-Loci refer to inferred K serotypes. Branch colours represent bootstrap support values.
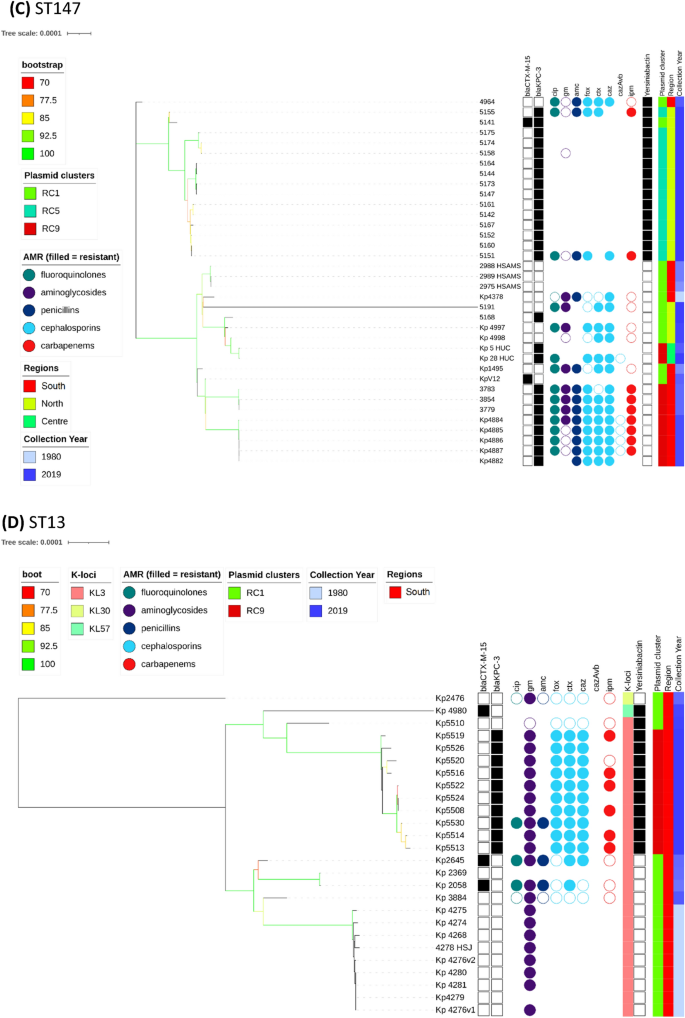
The most common ESBL (blaCTX-M-15) and carbapenemase (blaKPC-3) genes across the 509 Kp isolates by year group.
Most ST14 isolates (n = 20/26) fell within two dominant clades (Fig. 1B); the first clade (n = 11) was sourced from northern Portugal in year 2018 and contains the K16 locus, and the second (n = 9) was distinguished by the K2 serotype and spanned years 1980 to 2010. Only the second clade had blaKPC-3 carrying isolates (n = 6). Unlike for other STs, the genes used in phylogenetic reconstruction of ST14 were relatively concentrated within a 3.60 Mbp to 3.75 Mbp chromosomal region based on the NC_016845.1 Kp assembly (Fig. S1).
All ST147 (n = 36) isolates had KL64 and O2v1 serotypes and belonged to two main clades. One clade consisted of northern region isolates (n = 15) from year 2018, all of which carried blaKPC-3, but only one had blaCTX-M-15 (Fig. 1C). All isolates in this clade carried the siderophore yersiniabactin (ybt 16; ICEKp12) locus related to virulence. A second clade (n = 20) was sourced from the southern region and spanned years 1995 to 2016. Only 11 isolates in this clade carried blaKPC-3 and none had the yersiniabactin locus. Similarly, the ST13 tree had two clades characterised by the KL3 serotype (Fig. 1D). One clade (n = 10) was sourced from multiple hospitals in year 2019, and all isolates carried blaKPC-3 and yersiniabactin loci. Another clade (n = 13) spanned years 1982 to 2013 and came from the southern region. Only two isolates in this clade carried blaCTX-M-15, and none had other ESBL genes, blaKPC-3, or the yersiniabactin locus. Unlike the remaining ST13 isolates, these two phylogenetically distant isolates had inferred K57 and K30 serotypes.
Our dataset included four isolates of type ST1138, an ST which was previously described at two hospitals in Portugal13. We also had previously undescribed ST6003 (n = 3), which differs from ST1138 by an rpoB allele. All seven isolates were collected in 2012 from the same hospital in Lisbon. Remarkably, one of the ST1138 isolates has no AMR genes or mutations apart from blaSHV-36. This suggests its ancestors were newly introduced into the hospital system. This original strain has split into at least two lineages, both carrying blaKPC-3, but it is unclear if this acquisition was independent in each lineage. There are at least two inferred lineages because the isolates from overlapping dates have one of two types of rpoB alleles (denoted as alleles 1 or 46). We did not identify any post-2012 ST1138 isolates in our data, but it seems to have been present in Portuguese isolates collected in year 201113. Apart from ST6003, our dataset had two further one allele variants of ST1138: ST514 (n = 2) collected from hospital patients in the 1980s and ST323 (n = 1) collected from wastewater in 2018. Both ST514 and ST323 came from the same geographical area as sequence types ST1138 and ST6003.
Plasmid clustering
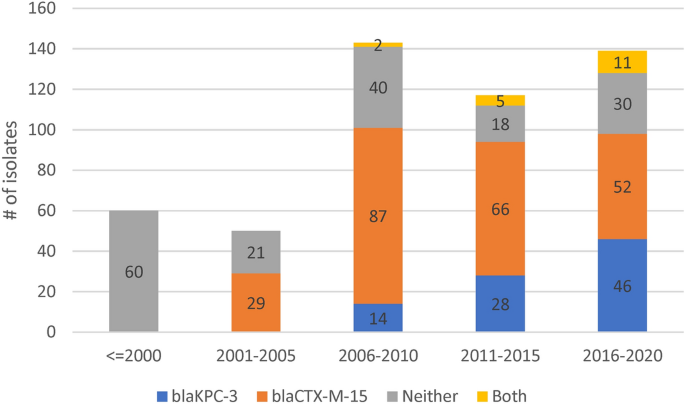
Of the 48 replicon families identified by PlasmidFinder software, six occurred in at least 10% of isolates (IncFIB 98%, IncFII 96%, IncFIA 55%, IncR 42%. IncHI1B 11%, IncN 10%). An average of 4 replicons (range: 0–9) were identified per isolate with only three isolates having none. To understand the pattern of plasmid replicons across isolates we applied UMAP dimensional reduction methods29 to presence-absence matrices of replicon family (n = 48) and unique replicon sequences (n = 240). The results based on families and exact replicon sequences were consistent; so here we present the higher resolution analysis using replicon sequences (Data S1). We observed nine clear and robust clusters (replicon clusters, RCs) (Fig. 3A). RC prevalence varied over time (Fig. 3B) with RC9, the cluster with 94% blaKPC-3 carriage, becoming dominant between years 2010 and 2019. By overlaying RC type onto the overall and ST specific phylogenetic trees, mosaic distributions were revealed, further supporting the movement of plasmids among Kp lineages (Fig. 1A–D).
(A) Clustering of isolates by their plasmid replicon (replicon clusters, RC) and antimicrobial resistance (AMR) genotypic profiles, revealing differentiation by carriage of blaKPC-3 and blaCTX-M-15 genes. X and Y axis are dimensions on which full data is projected, they are unitless; (B) Abundance of isolates from different plasmid clusters.
RC1 contained 57% of all isolates (n = 288), but very few (n = 9) isolates had blaKPC-3; although 59% carried ESBL encoding genes, which is consistent with early isolates dominating this cluster. The frequency of isolates in this cluster has declined substantially after year 2001. RC1 has a large diversity of plasmid replicons including IncFIB(K) (n = 214), IncFII(K) (n = 174), IncR (n = 108), and CoI(pHAD28) (n = 108) with each replicon family consisting of multiple distinct phylogenetic clades making RC1 interpretation complex. In contrast, the RC9 (n = 87) cluster has 25 different STs, including the dominant ST15, ST147 and ST14 types. Almost all Kp in RC9 cluster (94%, 82/87) carry blaKPC-3. This gene is absent in 5 isolates: ST14 from 2007 and 2010; ST416 from 2011; ST1138 from 2012; and ST359 from 2018. The key signature of RC9 is the presence of variants of FIA(pBK30683) and FII(pBK30683) replicons (Data S1). For isolates with blaKPC-3 in RC9, more than a third (31/87) were diverse STs collected between years 2010 and 2019, but possessing neither gyrA mutations, nor an aac(6′)-Ib-cr gene. These isolates were susceptible to fluoroquinolones with average inhibition zone diameters of 20 mm among 27 tested isolates.
The RC5 cluster (n = 14) consisted of ST147 carrying blaKPC-3 and was congruent with the clade sourced from northern Portugal in year 2018 (n = 15). Of these northern Portuguese isolates, 14 belonged to RC5 and one belonged to RC1 (Fig. 1C). The RC5 cluster is identifiable by two variants of IncN and IncFIB(pKPHS1) replicons (Data S1). IncN is uncommon in our dataset (51/2222 replicons) and its strong link with blaKPC-3 suggests, in line with earlier reports15,30, that blaKPC-3 may be mobilized by IncN plasmids. In contrast, other clusters had homogenous ST types: RC2, RC3, RC7 and RC8 consist of 57, 24, 11, and 8 ST15 isolates; RC6 consists of ten ST14 isolates; and RC4 of nine ST12 isolates. RC3 is interesting because despite having isolates from years 2003 to 2014, only one (1/24) had a blaSHV type beta-lactamase (blaSHV-11). These genes are very common (87.4%) and are normally chromosomal, which suggests either the loss of blaSHV or simultaneous circulation of several strains.
All RC2 isolates carried IncFIB(K) and IncFII(pKP91) replicons, where the associated variants were different to those from the RC1 replicons. The RC2 IncFIB(K) variant was also present in RC8 and RC7 isolates. RC7 isolates also carried a distinct variant of a Col440I replicon, and a variant of IncR is shared with RC1, RC2, RC3 and RC8. RC8 carried a distinct version of IncFII(K) and IncFIB(pQil) replicons. Finally, in addition to shared IncR and ColpVC variants, RC3 also carried IncHI1A and IncHI1B(R27) replicons, which were absent in other clusters (Data S1).
Antimicrobial resistance genotypes and phenotypes
In vitro AMR profiles and genotypes were analysed for the 509 Kp isolates, with some gaps depending on the decade of phenotypic assessment (Table S1). Most isolates underwent antibiotic susceptibility testing for aminoglycosides (n = 356, 68.1%), cephalosporins (n = 366, 71.2%), fluoroquinolones (n = 344, 66.9%), carbapenems (n = 296, 57.6%), and penicillins (n = 311, 60.5%) (Table S1). However, 121 isolates had no AMR susceptibility data. Since AMR testing was performed over multiple years, the concentration of active compounds in disks used might vary between isolates; all breakpoints were determined based on EUCAST v11.0 (2021)31.
Beta-lactams
The majority (75%) of 304 tested isolates showed susceptibility to imipenem; an antibiotic used widely in hospital clinical practice in Portugal (Fig. 4). The resistance driver was likely blaKPC-3 (n = 106), which was carried on a Tn4401d transposon in nearly all cases (n = 101/106). In four isolates [ST15 (Kp5149), ST147 (Kp5147), ST34 (Kp5148) and ST461 (Kp5162)], the blaKPC-3 gene has undergone an identical inversion within the Tn4401d structure. Only one of these four isolates (ST147) was tested for imipenem resistance and was determined to be resistant (inhibition zone diameter of 6 mm). This observation suggests the inversion did not significantly impair the resistance conferred by the presence of blaKPC-3. Additionally, our dataset contained three isolates without carbapenemase genes but resistant to imipenem. These three isolates had no clear commonality between them, nor clear distinction from susceptible isolates with a similar genotype.
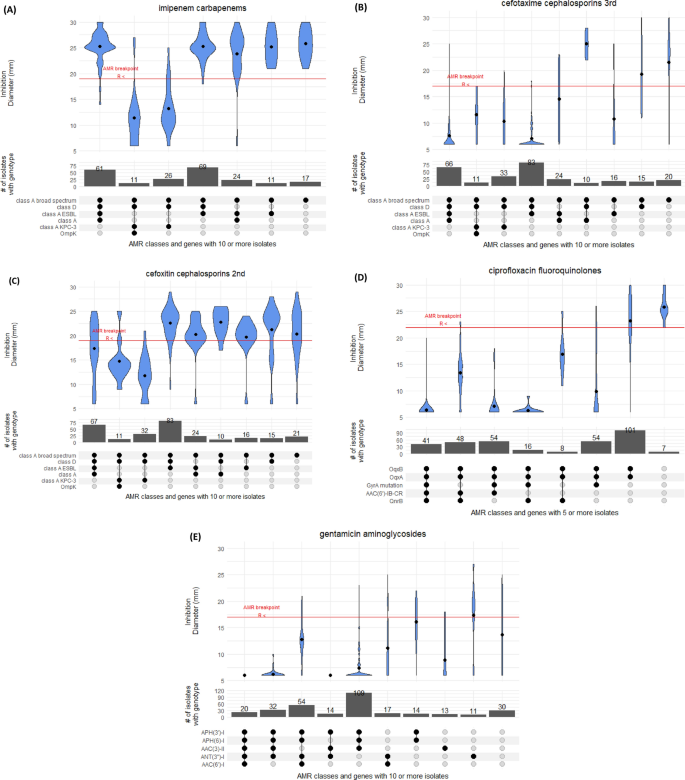
Distribution of inhibition zone diameters for different genotypes for (A) imipenem, (B) cefotaxime, (C) cefoxitin, (D) ciproflocaxin, and (E) gentamicin antimicrobials.
Resistance to cephalosporins was widespread with 68% of 335 tested isolates showing resistance to cefotaxime, a third-generation cephalosporin, driven likely by the presence of class A ESBL blaCTX-M-15 (n = 213, 41%) (Fig. 4). Ceftazidime had an even higher resistance prevalence of 91%. This drug was used widely in the 1990’s for Pseudomonas outbreaks, but is currently rarely used in monotherapies. The combination of ceftazidime and carbapenemase inhibitor avibactam (cazAvb) was rarely resistant (1/46 tests). The resistant isolate (ST15) carried blaOXA-1, blaTEM-1, blaSHV-28 and had truncated porin gene ompK36. Of the imipenem resistant isolates, twelve were tested for cazAvb and all were susceptible.
Finally, the second-generation cephalosporin cefoxitin presents an interesting case because its resistance profile was frequently inconsistent with its genomic or genotypic profile (Fig. 4). Of 366 tested isolates, 8 carried AmpC beta-lactamase blaDHA-1, which is associated with strong inhibition of cefoxitin. Consistent with this, these 8 isolates had a cefoxitin inhibition zone diameter of 6 mm. However, the inhibition zone diameter of the tested blaKPC-3 carrying isolates (n = 74) was on average 13 mm, which is below the 19 mm breakpoint, but well above the total inhibition diameter of 6 mm. In isolates with class A (non-broad-spectrum, broad-spectrum, ESBL) and D beta-lactamases, the susceptibility to cefoxitin varied vastly with isolates containing all four categories having almost uniform distribution of inhibition zone diameters between 25 and 6 mm. We were able to trace some of this inconsistency to 83 isolates with blaSHV-28. The mean diameter for isolates with class A broad spectrum, class A ESBL and class D genes is 22 mm. If this genotype also included blaSHV-28 (class A), the mean inhibition zone diameter reduced to 16 mm, but this effect is absent for other blaSHV variants.
Fluoroquinolones
Of the 307 isolates tested for ciprofloxacin susceptibility, 288 (94%) were resistant (Fig. S4). Five isolates did not have any common fluoroquinolone resistance determinants and were all susceptible with between 22 and 30 mm inhibition zone diameters. By far the strongest determinant of resistance was the presence of mutations in type II topoisomerase gyrA. In silico screening with Kleborate software identified simultaneous mutations GyrA83F and GyrA87A (n = 150), and single mutations GyrA83I (n = 91) and GyrA83Y (n = 15) as the most common ones. Nearly all isolates with gyrA mutations had inhibition zone diameters below 10 mm, whereas the resistance breakpoint is 22 mm. In 54 tested isolates that had both the gyrA mutation and qnrB gene, nearly all had inhibition zone diameters of 6 mm, demonstrating clear compounding of resistance. The oqxAB genotype gives a small decrease in susceptibility, but most of such isolates were still susceptible. Finally, qnrB and qnrS each substantially decreased inhibition zone diameter. In our dataset these genes were present only in combination with oqxAB, leading to reduced inhibition zone diameters of ~ 50% compared to oqxAB alone. As we did not have isolates with both qnr genes, we could not confirm if their effect is cumulative. Further, 36 and 24 isolates were tested with levofloxacin and norfloxacin, respectively, with 42% of each set showing susceptibility. In contrast to ciprofloxacin, levofloxacin was only moderately affected by gyrA mutations with an average inhibition zone diameter reduced to 14 mm (Fig. S4).
We examined the diversity of the parC gene which has been associated with fluoroquinolone resistance 5,32, and found 244 isolates carrying both the ParC80I mutation and gyrA mutations. The parC alleles were ST specific except for three ST307 outlying isolates that differ by a single SNP from the other seventeen ST307 and fifteen ST15 isolates (Fig. S5) which suggests a lack of selective pressure on the gene. A tree constructed using gyrA sequences formed clear clades for the abundant ST14 and ST15 types (Fig. S5). ST14 isolates formed two major clades, one linked to decades 1980’s and 2010’s (n = 14) and another from the 2000’s (n = 15). ST15 formed three clades, one linked to the 1980s (n = 13), and two others post-year-2000 (n = 18, n = 131). There was additional evidence of selective pressure in other STs for which we had fewer isolates (Fig. S5). We could not test the effect of parC mutations on resistance because all isolates with mutations in this gene also had mutations in gyrA. Finally, 237 isolates (46%) had an aac(6′)-Ib-cr gene, of which 132 were tested for ciprofloxacin resistance and 128 were resistant. This gene had moderate impact on disk diameter, but this impact is sufficient for isolates to fall below resistance threshold. While the majority of aac(6′)-Ib-cr carrying isolates also had gyrA mutations, we found 48 isolates with an oqxAB/qnrB/aac(6′)-Ib-cr genotype for which mean inhibition zone diameter was 13 mm compared to 17 mm for 8 isolates with an oqxAB/qnrB genotype.
Tetracycline
We tested Kp isolates for tetracycline (n = 57) and tigecycline (n = 48) resistance. While breakpoints for tetracycline are not standardised31, the results indicate very high resistance to the antibiotic (Fig. S4). The only five isolates susceptible to tetracycline were collected between years 1980 and 1982, which suggests in the remaining 52 isolates that resistance is acquired rather than intrinsic. Isolates with tetABD efflux pumps (n = 31/57) had marginally more resistant profiles. In contrast to tetracycline, most of tigecycline tests inhibition zone diameters were located just below the breakpoint, with only a few isolates with < 10 mm (Fig. S4). Kp does not have an EUCAST zone diameter breakpoint, so we used the E. coli 18 mm breakpoint instead. Interestingly, the tetABD efflux pumps did not reduce the inhibition zone diameter. Isolates without these efflux pumps were marginally more (mean of 14 mm versus 17 mm), not less, resistant.
Aminoglycosides
Among isolates tested with gentamicin (n = 349) and amikacin (n = 29), 89% and 55% were resistant, respectively. In those isolates that had no known gentamicin resistance determinants (n = 28), the average inhibition zone diameter was 15 mm versus an established breakpoint of 18 mm (Fig. 4). Two acetyltransferases were present in our isolates ((AAC(6’)-I, n = 283; AAC(3)-II, n = 291), of which AAC(3)-II was a stronger determinant of resistance, with nearly all inhibition zone diameters below 10 mm. AAC(6’)-I reduced the inhibition zone diameter from 15 to 12 mm, and the effect was cumulative with AAC(3)-II. Nucleotidyltransferase ANT(3’’)-I and phosphatases APH(6’)-I and APH(3’)-I, even combined, appear to have a marginal effect on resistance. The 16S rRNA methyltransferase gene armA was present in two isolates, and the gene rmtB was present in two additional isolates.
Virulence determinants
Using VFDB, Kleborate and BLAST tools, we examined the dataset for virulence genes33,34,35. The type 1 fimbriae locus, fimABCDH, was present in nearly all isolates (501/509, 98%). It was absent in isolates from ST960 (4/4), ST15 (3/161) and ST76 (1/3). Similarly, the type 3 fimbriae locus, mrkABCDF, was present in all isolates except for one (ST147). In contrast, iron uptake locus kfu was present in 245 (50%) isolates. This locus was perfectly correlated with ST; no ST had simultaneously kfu positive and negative isolates. Among the most frequent STs, ST15 (n = 161), ST13 (n = 26) and ST14 (n = 26) isolates had a kfu locus, but it was absent in ST147 (n = 36), ST348 (n = 22), and ST307 (n = 20) isolates. There were only a few uncommon virulence genes. For example, we did not find any rmpA and rmpA2 regulators of hypermucoviscosity. Two isolates (Kp4248 and KpV9) had aerobactin siderophores: one with iucA2 (ST3) and the other with iuc3 (ST3027). Both isolates had limited known AMR determinants. ST3 had only blaSHV-1 and ST3027 had APH(6’)-Ia/d, tet(A) and blaSHV-33. The salmochelin locus iroBCDEN was found only in some ST48 isolates (n = 7/9), all of which carried ESBL blaCTX-M-15. Of these 7 isolates, 6 came from the same Lisbon hospital between years 2005 and 2009. Yersiniabactin was present in 56% of isolates, and was carried most frequently on integrative conjugative elements ICEKp3, ICEKp4 and ICEKp12 at 21%, 15% and 6% of all isolates, respectively. Additional screening against the VFDB virulence database only revealed astA and cseA genes in one isolate.
Genomic epidemiological analysis of Klebsiella pneumoniae from Portuguese hospitals reveals insights into circulating antimicrobial resistance & Latest News Update
Genomic epidemiological analysis of Klebsiella pneumoniae from Portuguese hospitals reveals insights into circulating antimicrobial resistance & More Live News
All this news that I have made and shared for you people, you will like it very much and in it we keep bringing topics for you people like every time so that you keep getting news information like trending topics and you It is our goal to be able to get
all kinds of news without going through us so that we can reach you the latest and best news for free so that you can move ahead further by getting the information of that news together with you. Later on, we will continue
to give information about more today world news update types of latest news through posts on our website so that you always keep moving forward in that news and whatever kind of information will be there, it will definitely be conveyed to you people.
Genomic epidemiological analysis of Klebsiella pneumoniae from Portuguese hospitals reveals insights into circulating antimicrobial resistance & More News Today
All this news that I have brought up to you or will be the most different and best news that you people are not going to get anywhere, along with the information Trending News, Breaking News, Health News, Science News, Sports News, Entertainment News, Technology News, Business News, World News of this made available to all of you so that you are always connected with the news, stay ahead in the matter and keep getting today news all types of news for free till today so that you can get the news by getting it. Always take two steps forward
Credit Goes To News Website – This Original Content Owner News Website . This Is Not My Content So If You Want To Read Original Content You Can Follow Below Links
